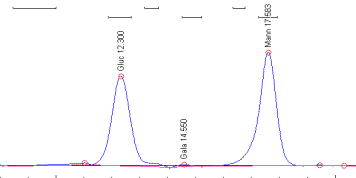
Student Research Projects from Spring 2005.
In 1999 our faculty began asking the question: Are undergraduate science laboratories teaching students the art and trade of science or simply leaving them with a memory
of trivial experiments done for unknown reasons? From our conversations with students in biology, it certainly appeared as though the latter was the consensus. Students
used words like: "boring", "restrictive", "pointless", and so on, to describe the biology laboratory. In fact, very few of our students characterized the lab to be a good
learning experience. Even our 'best and brightest' students agreed that while our new cutting-edge DNA genomics labs were fun, structured labs really didn't help them
learn. In fact, they indicated that they often didn't really understand what they were doing until the week after completing the experiment, when they wrote the lab report.
In an effort to remedy this, we began a long-term redesign of the biology sequence in the Lyman Briggs School of Science at Michigan State University. Combining what
educational experts have found about active and cooperative learning and challenging our own biology faculty to make the lab as realistic as possible, the lab curriculum
departed from numerous 3-hour traditional labs that each student performed on their own, to what we now term "Teams and Streams." Now we use student research teams to pose
a scientific question/hypothesis, propose an experimental design to set about gathering evidence for support of said hypothesis, perform multi-week investigations and then
present their findings in various forms (web sites, interviews, and multiple drafts of a scientific manuscript along the way).
Stream II Cellular Physiology Stream:
For example, in Stream I (the Cell Physiology labs), student research teams work for 7 weeks to produce and submit their final manuscript. In the first week they are introduced to the lab and it is
then that they develop their research question(s) and plan. While they are then trained in various lab techniques and assays during weeks 2, 4, and 6 (by performing very
structured traditional labs), during weeks 3, 5, and 7 groups pursue their independent research projects by re-applying and extending the range of different assays they
learned in structured labs to answer their own questions in their independent investigations.
The groups each have a designated space in the laboratory classroom (a lab bench) that is fully equipped with computers and scientific equipment to help them in their novel
studies. In addition, students are allowed to return to lab as often as they wish to complete their research --they reserve open benches online. Since they create their own
methods, we find students can better explain how and why the equipment is used to complete their project. The response to this classroom laboratory design has been
overwhelmingly positive. As students once dreaded the 3 hours per week that they were in lab, we now find them signing up for additional lab time to where they are even
exceeding 10 hours/week in lab! Not only are these students getting a real taste for what science really is, but they are finding, even in this short time, how much they
enjoy it.
----- Sample Student websites reporting their findings at the end of Stream I independent investigations -----
Team "Eagles": Effects of Citrus Fruits as Fertilizers Using Photosynthetic Testing, Growth Rate and Leaf Area Ratio of Hedera helix
Average Joes: Enzyme production is relocated and photosynthesis becomes more efficient as Nepenthes tissues mature.
The Flying Hellfish: Sugar, Photosynthesis, and pH Tests Reveal Inhibitory Effect of Acid Rain on Hedera helix L.
3 Girls and a Guy: Carbohydrate, Protein, and Photosynthesis Levels Show Inconclusive Results on Maintaining Rosa multiflora Health.
The Green Mile: Sugar, action spectra, and protein/phosphate study of variants of Brassica oleracea yields similarity.
Group Guinness: Sugar, Starch, Photosynthetic Pigments, and Protein Comparison in Musa acuminata (Banana) Ripening Stages
Katkus: Pigments, Reducing Sugar Richness & Structure, & Protein Richness Varied Due to Cooking & Freezing Brassica oleracea
Krazy Kangaroos: Three species of Lycopersicon esculentum differ in sugar and protein content after sugar, HPLC, protein and pigment tests.
The Luckie Charms: Carbohydrate, Bacteria, and Various Other Tests Reveal Only Minute Differences In Organic and Conventionally Grown Fruits
Nalgene: Brassica oleracae , the Low-Carb Alternative for Solanum tuberosum based on Barfoeds, Iodine and Bradford assays.
Orange County: Energy drinks found as a better source of energy than teas by testing sugar, enzyme, protein and caffeine levels
Purple Cobras: Herbal Supplements Lead to Better Test Results in Photosynthesis and Regeneration in Fatshedera lizei
The Spartanettes: DEGRADING STRUCTURAL EFFECTS SEEN IN GATORADE¬ TREATED BRASSICA RAPA, ANALYZED BY SUGAR, PROTEIN AND ACTION TESTS.
Team 19: Carbohydrate and Protein Content Varies Moderately In Dried Bananas, Peaches, and Cranberries Compared to Fresh.
Team Fusariums: Loss Of Photosynthetic Pigments, Vitamin C, and Sugars in Brussel Sprouts Cooked at Various Times
Team Gandalf: Fertilizer, Pesticides and Nutrient effects on Brassica Rapa.
Team Science: Iodine Test Reveals CaCl2 Up to 0.0676M Enhance Amylase Digestion of Solanum tuberosum Starch
The Amoebas: Carbohydrate and Protein Assays Show Soy and Whey Protein Supplements are Advantageous in Improving Overall Health .
The Carbs: Analysis of Festuca rubra using Bradford, Hills, Height and Sugar Tests to Determine the Effects of Added Fertilizer
The Cells: Carbohydrate, Protein, Photosynthetic Pigments, & Caffeine Assays Show Variation: Green vs Roasted Coffea arabica
The Eggs: Grade A Chicken Eggs vs. Eggbeaters: sugar, protein, photosynthesis, lipid tests find differences in lipid quantity
Team Vitamin C: Sugar, Protein, and Vitamin C Tests Reveal Organic Capsicum annum is More Nutritious than Genetically Modified
Vortex Genies: Photosynthetic and Sugar Analysis of Green Beans (Phaseolus vulgaris) Confirm EDTA as Best Preservative
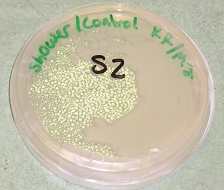
Stream II Molecular Physiology or DNA Stream:
Students were
required be much more independent in Stream 2 (the final 7 week DNA or Molecular Physiology investigation) than in the previous stream.
They were provided
with:
(i) series of primers: 1-page explanations of plasmids, antibiotics etc,
(ii) a section containing info on genomic isolation and PCR
techniques (compilation of protocols and review articles published elsewhere),
(iii) a protocol
section that was taken directly from a laboratory protocol manual entitled Molecular
Cloning by Maniatis et al (1989), and a Stream 2 appendix.
Their instructions indicated: "The Molecular Cloning
manual is used both in the academic and industrial settings by scientists all over the
world as a resource for standard protocols in molecular biology. The original Molecular
Cloning manual has been distilled down to what we will call "mini-Maniatis." This
section along with "PCR Information" section (located prior to mini-Maniatis) contains
all the specifics that will assist you in completing your goals. In fulfilling the Stream II
of laboratory your group will have three options for your independent investigations. You
may either choose to (i) develop a "Genome Fingerprint Assay" for your specimens
from Stream I, (ii) pursue a "Plasmid Identification Inquiry" of naturally occurring
antibiotic resistant bacteria, or (iii) design your own independent research project of another topic."
----- Student Groups and Stream II Research -----
By: Nathan Anthony Althaver: Extraction of Musa acuminate DNA and PCR using Rapid Primers
By: Lubna Ansari: FAILED ATTEMPTS TO IDENTIFY A BACTERIAL ANTIBIOTIC RESISTANT PLASMID FROM BATHROOMS USING GEL ELECTROPHORESIS.
By: Nidhi Atri: Searching at "Holmes" for an Antibiotic-Resistant Plasmid by Isolation, Amplification, and Purification
.
By: Bradley Damm: Comparing Antibiotic Resistant Bacteria Found on a Human and Objects of Contact by Lysis and Mapping of Plasmids.
By: Jessica Grover: Kanamycin Resistant Bacteria in Red Cedar Water; Gel Electrophoresis and Restriction Digest Lead to Unknown Plasmid.
By: Jason Harner: Comparison of the location of antibiotic resistant bacteria by the identification of plasmids using gel electrophoresis
By: Nicole Hedquist: DNA Extracted from Human Hair Follicles gave Inconclusive Fingerprints after using AP-PCR and Gel Electrophoresis
By: Jasmine Hill: Antibiotic-Resistant Bacteria Found That Contains Plasmids Which Are Identified Using Gel Electrophoresis.
By: Rebecca Hurst: ATTEMPTED IDENTIFICATION OF AN ANTIBIOTIC RESISTANT BACTERIAL PLASMID USING GEL ELECTROPHORESIS
By: Jeannette Kelly: Restriction Digest Unable to Identify Plasmid in Antibiotic-Resistant Bacteria Using Gel Electrophoresis.
By: Devin Murphy: in Attempt to Locate
By: Kevin Ogden: Triclosan Promotes Kanamycin Resistance: Characterizing a Plasmid by Restriction Digestion and Electrophoresis
By: Sonia Rahangdale: The Use of Restriction Digest & Gel Electrophoresis to Identify a Plasmid Conferring Ampicillin Resistance to Bacteria.
By April Roodbeen: Human Hair Extraction and Amplification through AP-PCR Procedure Produces Inconclusive DNA Fingerprint.
By: Eric M Rosenbaum: Genomic Fingerprint Assay for Emerald Ash Borer Larvae fails using primers DL7-10 in AP PCR and Gel Electrophoresis
By: Samantha Salvia: Plasmid in antibiotic-resistant bacteria isolated by lysis, cut by restriction enzymes, gauged by gel electrophoresis.
By: Marisa Solano-Sanchez: The Search for the Emerald Ash Borer Genomic Fingerprint by means of PCR and Gel Electrophoresis
By: Paul Tomaszycki: Bacteria Do Not Need R-plasmids In Order To Be Resistant To Antibiotics.
By: Jillian K. Trombley: RAPD PCR using Primers DL7, DL8, DL9, DL10 and Gel Electrophoresis with Emerald Ash Borer for Genomic Fingerprinting
By: Dustin Wayo: Identifying Kanamycin Resistant Plasmids Using Restriction Enzymes and Gel Electrophoresis
By: Zimin Zhao: Identification of Plasmid from Antibiotic Resistant Bacteria by Amplification, Restriction Enzyme Digestion and Gel Electrophoresis

